Research
Our Science
Q: How is our brain shaped by our experiences?
Everything we experience produces electrical activity in our brain. Brain activity turns on a particular set of genes in each different kind of brain cell that allow us to adapt to and learn from our environment. This neuronal activity-dependent gene expression is essential for healthy brain development, function, and maturation. While the transcriptional and epigenetic effects of neuronal activity have been systematically explored in rodents, very little is known about these adaptive changes in human brain cells.
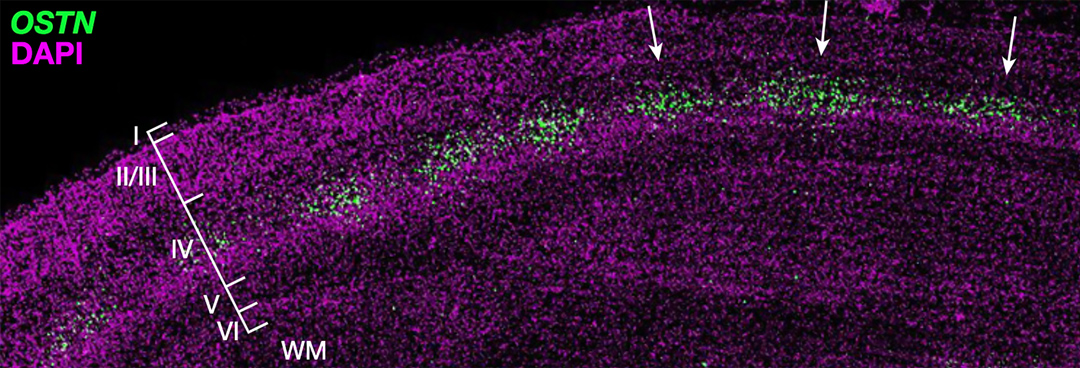
OSTN expression is induced by sensory experience in the primate cortex.
In human neurons in vitro, and in non-human primates in vivo, the Boulting Laboratory investigates activity-dependent transcriptional and epigenomic dynamics that cannot be studied in other animal model systems. By identifying novel patterns of gene regulation, we demonstrate how these critical gene expression programs have evolved to vary between species. We have uncovered both primate-specific neuronal activity-regulated genes, as well as evolutionarily-conserved genes that have been repurposed in the primate brain. Ongoing experiments explore how the molecular functions these genes impart new adaptive functions to the human brain.
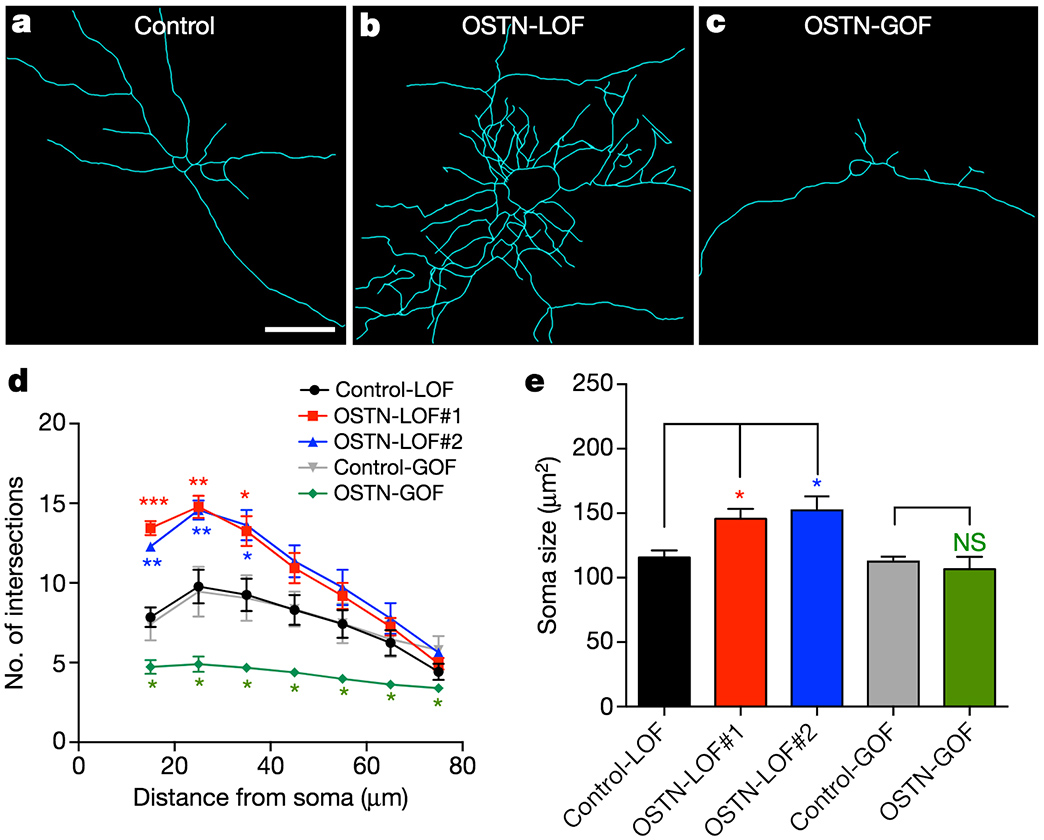
OSTN regulates activity-dependent dendritic growth in human neurons.
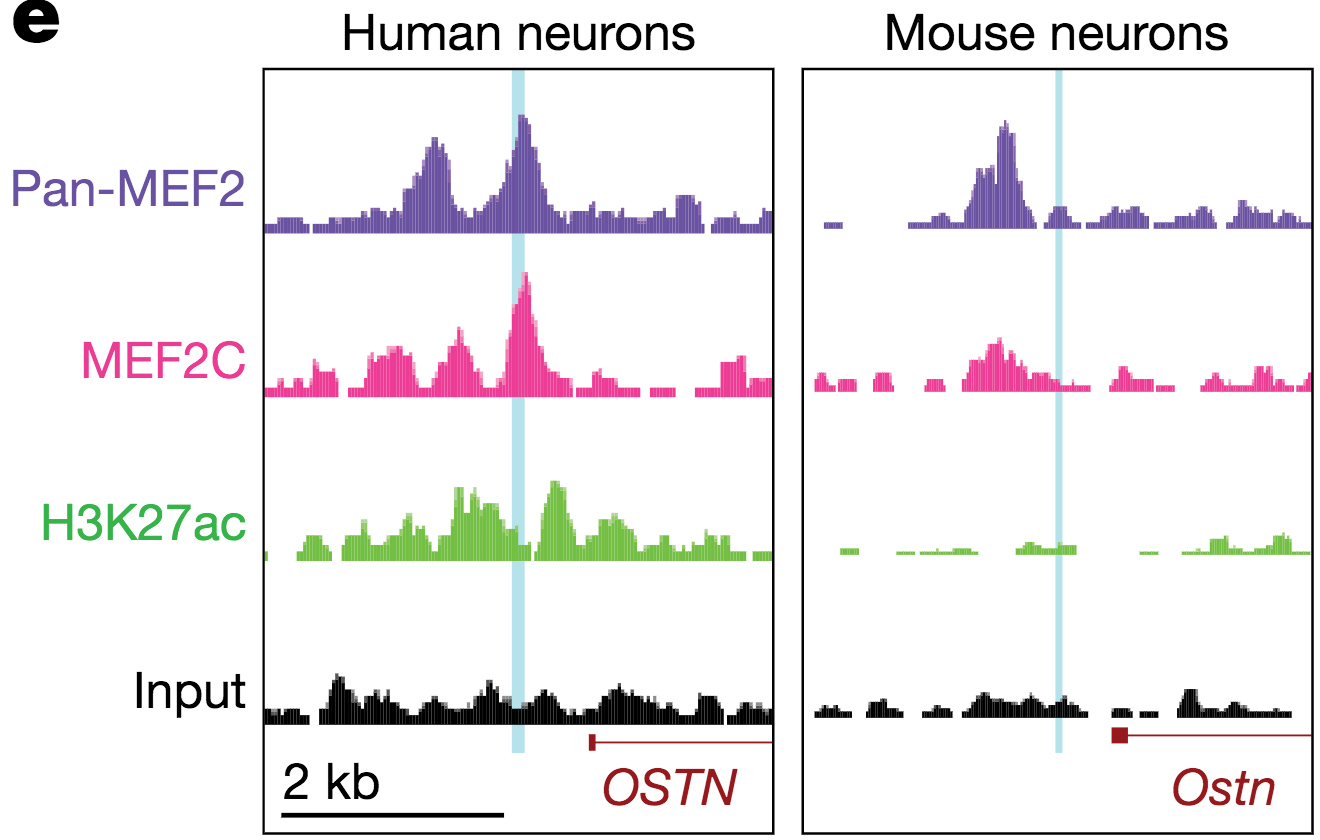
Primate-specific enhancer regulation by MEF2 drives neuronal activity-dependent induction of OSTN.
One example is the gene Osteocrin (OSTN) that encodes a small secreted peptide. Dr. Boulting helped identify OSTN as selectively induced by neuronal activity in human and macaque brain, but not that of mouse or rat. Previous studies showed that the induction of OSTN in primates relies on a series of base-pair changes within an otherwise conserved enhancer sequence. And that OSTN expression dramatically regulates dendritic outgrowth of developing human neurons.
The Boulting lab is now continuing its investigation into the regulatory evolution and function of OSTN, and the transcription factors that control its expression (SATB2 and MEF2C), in order to uncover new aspects of human neuronal gene regulation during brain development and disease.
Q: What makes the human brain unique?
The human brain is enormous and complex, with an estimated 86 billion neurons, expanded cellular diversity, increased neuronal connectivity, and protracted stages of development compared to other mammals1–4. These differences are thought to contribute to human-specific characteristics such as refined digital dexterity, symbolic language, and abstract thought2. How the evolutionary changes in our brain’s development and adaptive capacity have given rise to human or even primate-specific characteristics remains largely uncharacterized.
The development and maturation of the brain is dependent on neuronal activity in a wide range of animal species5. Because human brain structures continue to mature over an extraordinarily long timeline of 20-30 years, it seems we are one of the few animals in which sensory input continuously shapes and refines neural circuit architecture over decades.
The limited availability of primary human brain tissue in which neuronal activity can be manipulated has largely precluded molecular characterization of these responses in humans. The Boulting Laboratory uses human stem cells to generate particular populations of human neurons in order to discover the molecular genetics of human neuronal development, cellular identity, and neuronal activity-dependent gene expression.
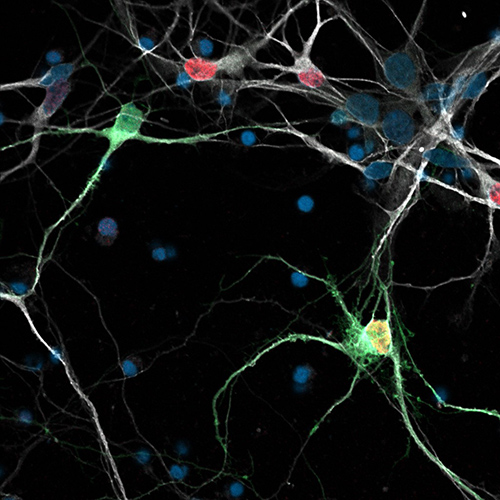
Human GABAergic stem cell derived neurons growing in culture.
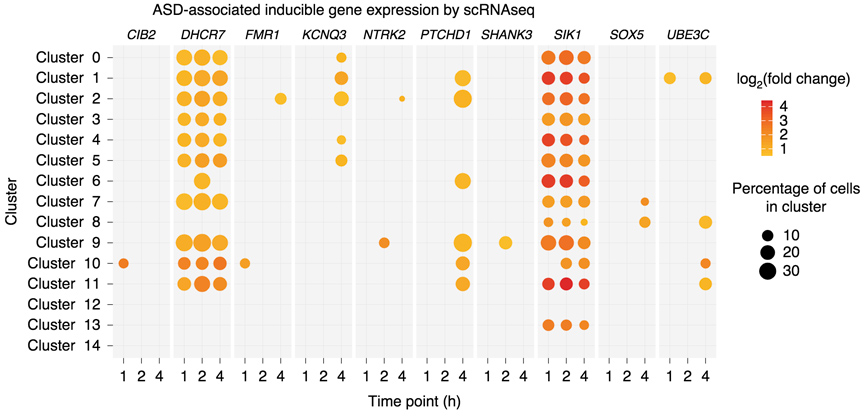
Neuronal activity-dependent gene expression of autism associated genes in human GABAergic stem cell derived neurons, measured by single-cell RNA sequencing.
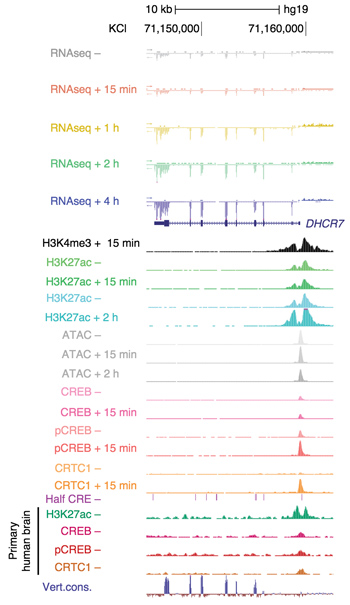
Multiomic view of the evolutionarily- divergent promoter of the autism-associated gene DHCR7 in human GABAergic stem cell derived neurons.
2. Sousa AMMM, Meyer KA, Santpere G, Gulden FO & Sestan N Evolution of the Human Nervous System Function, Structure, and Development. Cell 170, 226–247 (2017). [PMC free article] [PubMed] [Google Scholar]
3. Lui JH, Hansen DV & Kriegstein AR Development and Evolution of the Human Neocortex. Cell 146, 18–36 (2011). [PMC free article] [PubMed] [Google Scholar]
4. Petanjek Z et al. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proceedings of the National Academy of Sciences 108, 13281–13286 (2011). [PMC free article] [PubMed] [Google Scholar]
5. Hensch TK Critical period regulation. Annual review of neuroscience 27, 549–79 (2004). [PubMed] [Google Scholar]
Q: How does our primate-evolved genetic code contribute to neurodevelopmental and psychiatric disease?
GABAergic neuronal circuits are implicated in several neurological disorders. In Dr. Boulting’s postdoctoral work, she conducted a comprehensive activity-dependent transcriptional and epigenetic profiling of human induced pluripotent stem cell (hiPSC)-derived GABAergic neurons similar to those of the early developing striatum. This study identified genes whose expression is inducible following membrane depolarization, some of which have specifically evolved in primates, and/or are associated with neurological diseases, including schizophrenia, bipolar disorder, and autism spectrum disorder (ASD).
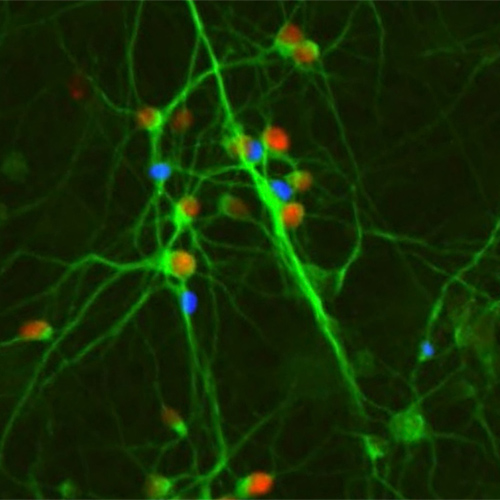
Human cortical neurons expressing the disease-associated transcription factor protein SATB2 (red nuclei), derived from either human pluripotent stem cells in vitro (above left) , or from primary human brain tissue (above right).
This finding, that depolarization-induced genes of human GABAergic cells are enriched for ASD-association, taken together with the neurological disease heritability enrichment observed in activity-responsive gene regulatory elements, suggests a role for dysregulated calcium-dependent gene expression within developing striatal neurons in ASD etiology.
One of our current research directions focuses on how the primate and human brain activity-dependent transcriptional and epigenetic response may be different from other animals, and how subtle genetic changes can lead to important and disease-relevant differences in human brain cells. We hope this will enable improved fine mapping of neurological disease-associated genome sequence variants within neuronal activity-dependent regulatory elements and lay the foundation for future comparisons between the activity-dependent regulatory landscapes of different human neuronal subtypes.
Graduate Student Rotation Projects
Graduate students who rotate in our lab will each be assigned participation in a specific project, like one of the sample projects summarized here.
Example # 1 :
Determinants of human neuronal transcriptional specification
We found that the conserved gene Osteocrin (OSTN), was specifically expressed in subsets of human and primate neurons, but not in rodents, and is likely to control neuronal connections in response to brain activity. Understanding how “non-coding” genetic sequences evolved to direct this new feature of the primate brain is an important step toward uncovering the genetic logic that determines how our brain is built. The goals of this project are to identify primate-evolved regulatory sequences controlling the OSTN gene while determining the human genome-wide binding patterns of psychiatric disease-associated neuronal transcription factors. This project includes human stem cells, genomic biochemistry, and bioinformatics.
Example # 2 :
Neuronal activity-dependent transcription of the primate brain.
Neuronal activity-dependent gene expression is required for proper for neuronal differentiation, circuit formation, behavior and learning. While the transcriptional and epigenetic effects of neuronal activity have been systematically explored in rodents, our work has shown important differences in this critical pathway between rodents and primates, and dozens if not hundreds more novel activity-dependent gene expression patterns have yet to be uncovered. SHARE-seq is an emerging technology from the Buentrostro Lab at Harvard University that collects both ATAC-seq and RNA-seq data from each individual nucleus. In this project, SHARE-seq is used to simultaneously identify cell type-specific activity-regulated transcripts, and the DNA regulatory elements that control them, in unique samples from macaque primary visual cortex. This project includes next-generation sequencing, single-cell multiomics, and bioinformatics.
Example # 3 :
OSTN function in developing human cortical neurons
Why has OSTN evolved to be expressed in such a specific pattern in the human brain? The answer lies in figuring out what OSTN helps those neurons achieve. This project uses human stem cell lines with mutated OSTN alleles to understand the effects of OSTN loss of function on differentiation, transcriptional profiles, and neuronal morphology. Because we previously showed that OSTN affects dendritic arborization, which is a determinant of neuronal connectivity, we will also use electrophysiology to explore neuronal activity characteristics and synaptic phenotypes between the human neurons of different OSTN genotypes in vitro and in vivo though xenotransplantation into the rodent brain.